42 label each structure in the following diagram of mrna processing. not all labels will be used.
No, because some eukaryotic genes have introns that are not translated. ... Label each structure in the diagram of mRNA processing. Not all labels will be ... Problem: Label each structure in the following diagram of mRNA processing. Not all labels will be used. FREE Expert Solution.
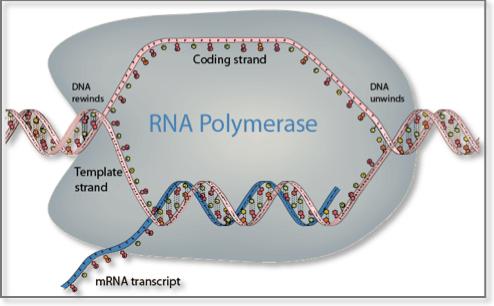
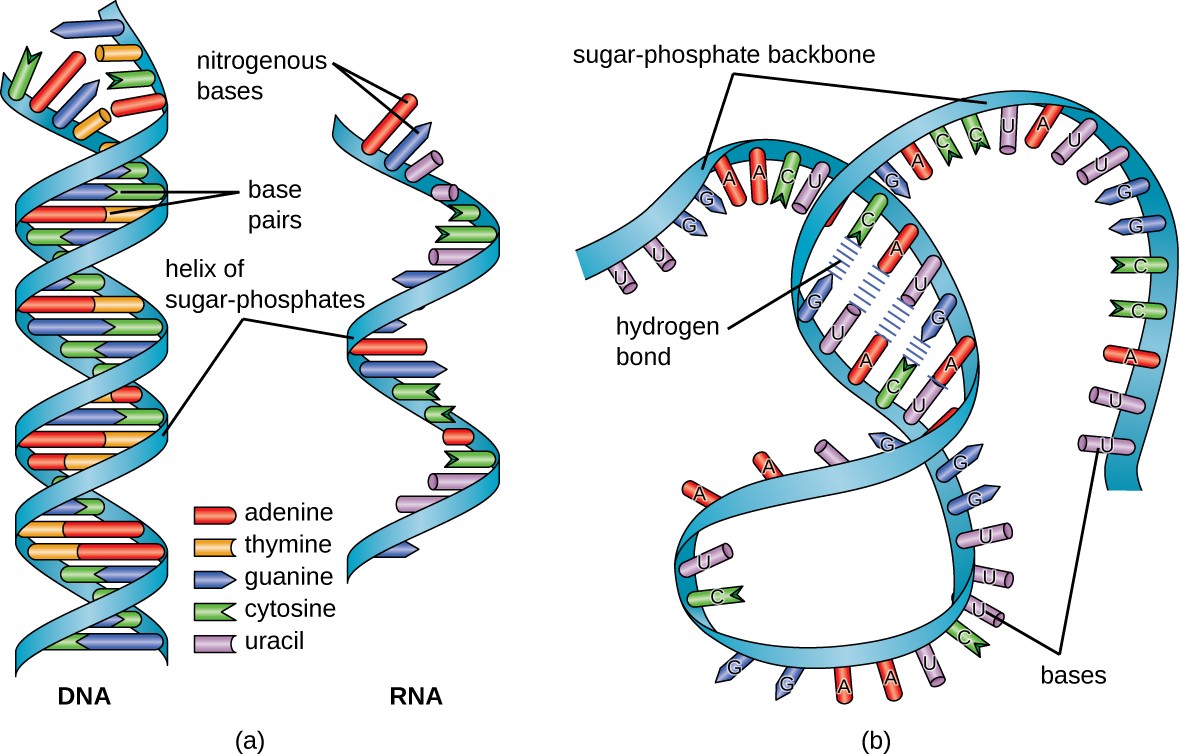
Messenger RNA (mRNA) =. Messenger RNA (mRNA) is a single-stranded RNA molecule that is complementary to one of the DNA strands of a gene. The mRNA is an RNA version of the gene that leaves the cell nucleus and moves to the cytoplasm where proteins are made. During protein synthesis, an organelle called a ribosome moves along the mRNA, reads its base sequence, and uses the genetic code to translate each three-base triplet, or codon, into its corresponding amino acid.

Label each structure in the following diagram of mrna processing. not all labels will be used.
Label each structure in the following diagram of mRNA processing. Not all labels will be used. Questions 11-14 Use asnwers A – E below pre-mRNA 1 30 31 104 ... Experts are tested by Chegg as specialists in their subject area. We review their content and use your feedback to keep the quality high. 1- Exon …. View the full answer. Transcribed image text: Label each structure in the following diagram of mRNA processing. Not all labels will be used. The secondary structure of a typical tRNA, in this case tRNAAla, is shown in Figure 1, below. The structure consists of hydrogen bonded stemsand associated loops, which often contain nucleotides with modified bases (e.g. inosine[I], ribothymidine [T], pseudouridine[Ψ], methylguanosine[D]). Figure 1. Schematic of tRNA (tRNAAlanine) secondary structure. Figure modified from Becker, et al., The World of the Cell. The tertiary structure of all tRNAs is similar to that of tRNAPhe, at left, a canonical "L-shaped" molecule. As can be seen, the "cloverleaf" secondary structure shown in Figure 1 results in a complex three dimensional folding of the molecule. The amino acid attachment site at the 3' end and the anticodon loopare observed at the two ends of the "L." The hydrogen bonded stems stabilize the tertiary structure. The Acceptor and Anticodon stemsare indicated. Modified bases in the Anticodon, T, and Dloops are indicated by thick wireframe. The anticodon bases project from the antico...
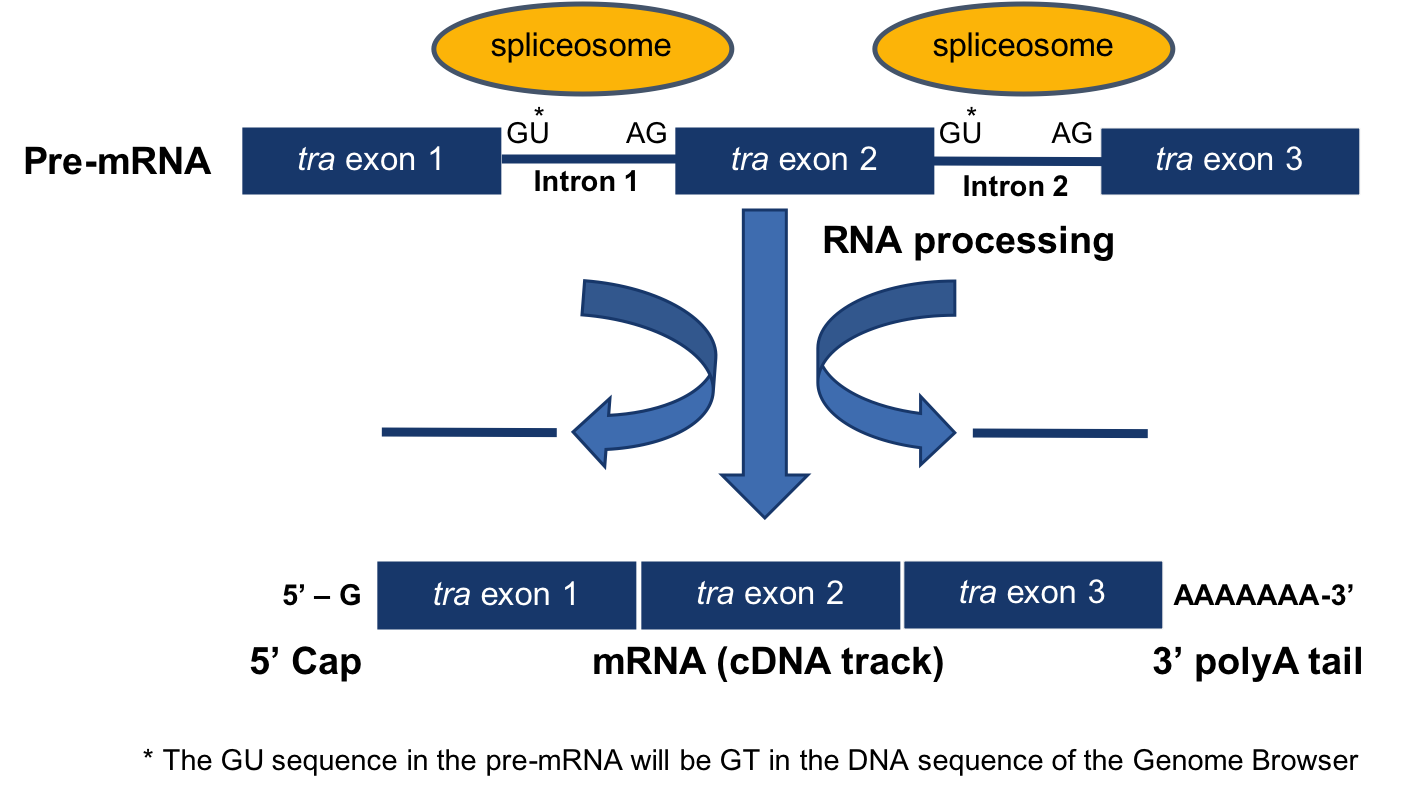
Label each structure in the following diagram of mrna processing. not all labels will be used.. mRNA Processing • Cap –7-methyl guanosine (7mG) added to 5’ end of mRNA –Protects against nucleases –Recognition for transport out of nucleus • Tail –Poly-A sequence added to 3’ end of mRNA –3’ terminal end cleaved at AAUAAA sequence before polyadenylation (most mRNA) –Protects from degradation The mRNA carries the message from the DNA, which controls all of the cellular activities in a cell. If a cell requires a certain protein to be synthesized, the gene for this product is “turned on” and the mRNA is synthesized through the process of transcription (see RNA Transcription). Label each structure in the following diagram of mRNA processing. Not all labels will be used. prr-mKNA I , p rt- m R NA mRNA ')3. Coding •c gmcn: Label each structure in the following diagram of mRNA processing. Not all labels will be used. Questions 11-14 Use asnwers A - E below pre-mRNA 1 30 31 104 ...1 answer · 0 votes: Answer:-
Biology. Biology questions and answers. Label each structure in the following diagram of mRNA processing. Not all labels will be used. Questions 11-14 Use asnwers A - E below pre-mRNA 1 30 31 104 105 146 + 1 12.. pre-mRNA 30 31 105 146 13.. Coding segment 14.. mRNA 146 exon 5-cap polymerase poly-A tail intron. The secondary structure of a typical tRNA, in this case tRNAAla, is shown in Figure 1, below. The structure consists of hydrogen bonded stemsand associated loops, which often contain nucleotides with modified bases (e.g. inosine[I], ribothymidine [T], pseudouridine[Ψ], methylguanosine[D]). Figure 1. Schematic of tRNA (tRNAAlanine) secondary structure. Figure modified from Becker, et al., The World of the Cell. The tertiary structure of all tRNAs is similar to that of tRNAPhe, at left, a canonical "L-shaped" molecule. As can be seen, the "cloverleaf" secondary structure shown in Figure 1 results in a complex three dimensional folding of the molecule. The amino acid attachment site at the 3' end and the anticodon loopare observed at the two ends of the "L." The hydrogen bonded stems stabilize the tertiary structure. The Acceptor and Anticodon stemsare indicated. Modified bases in the Anticodon, T, and Dloops are indicated by thick wireframe. The anticodon bases project from the antico... Experts are tested by Chegg as specialists in their subject area. We review their content and use your feedback to keep the quality high. 1- Exon …. View the full answer. Transcribed image text: Label each structure in the following diagram of mRNA processing. Not all labels will be used. Label each structure in the following diagram of mRNA processing. Not all labels will be used. Questions 11-14 Use asnwers A – E below pre-mRNA 1 30 31 104 ...

Identification Of The Cellular Targets Of The Transcription Factor Tcerg1 Reveals A Prevalent Role In Mrna Processing Journal Of Biological Chemistry
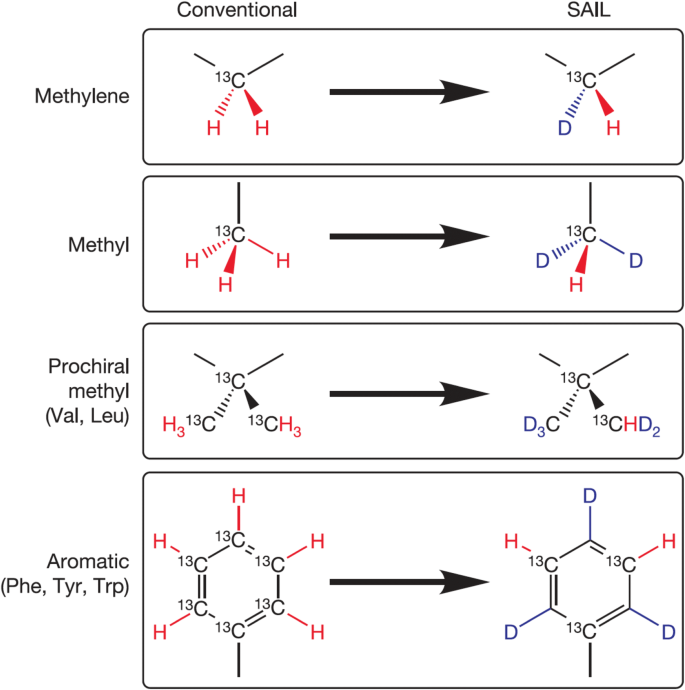
Accessing Structure Dynamics And Function Of Biological Macromolecules By Nmr Through Advances In Isotope Labeling Springerlink
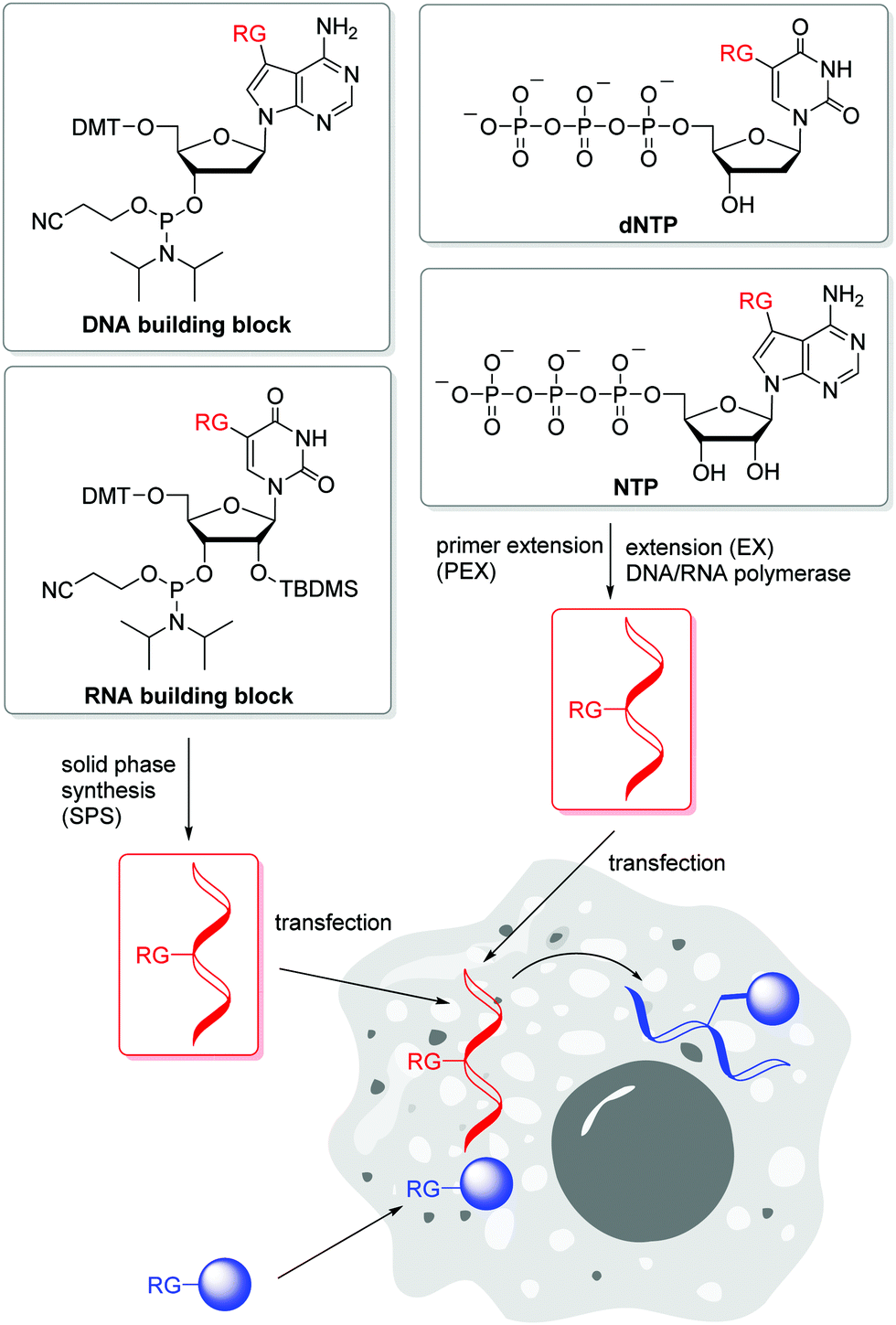
Labelling Of Dna And Rna In The Cellular Environment By Means Of Bioorthogonal Cycloaddition Chemistry Rsc Chemical Biology Rsc Publishing Doi 10 1039 D0cb00047g
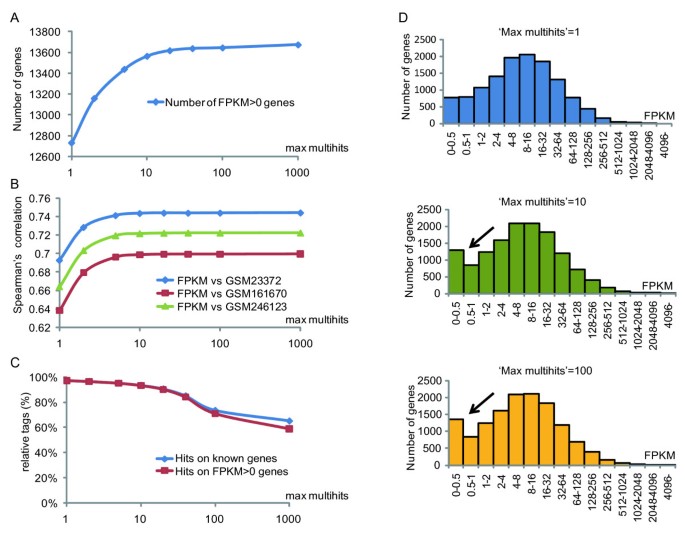
The Classification Of Mrna Expression Levels By The Phosphorylation State Of Rnapii Ctd Based On A Combined Genome Wide Approach Bmc Genomics Full Text

Peripheral Insulin Regulates A Broad Network Of Gene Expression In Hypothalamus Hippocampus And Nucleus Accumbens Diabetes

32 Label Each Structure In The Following Diagram Of Mrna Processing Not All Labels Will Be Used Labels Design Ideas 2020

Metabolic Labeling And Imaging Of Cellular Rna Via Bioorthogonal Cyclopropene Tetrazine Ligation Ccs Chem

Conserved Long Range Base Pairings Are Associated With Pre Mrna Processing Of Human Genes Nature Communications
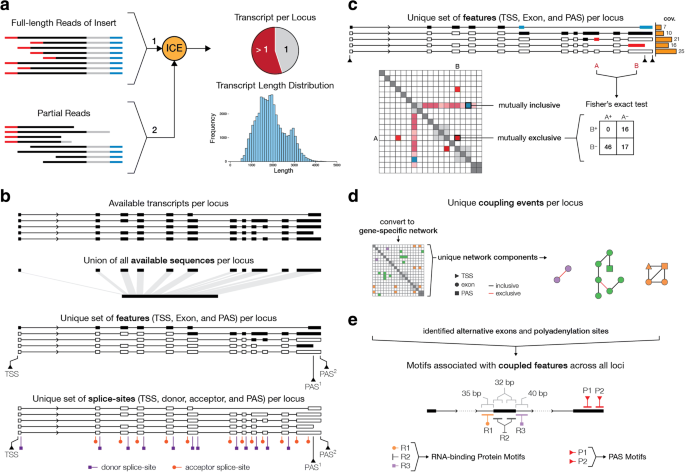
Full Length Mrna Sequencing Uncovers A Widespread Coupling Between Transcription Initiation And Mrna Processing Genome Biology Full Text

Progress And Challenges In Mass Spectrometry Based Analysis Of Antibody Repertoires Trends In Biotechnology

Proximity Rna Labeling By Apex Seq Reveals The Organization Of Translation Initiation Complexes And Repressive Rna Granules Sciencedirect
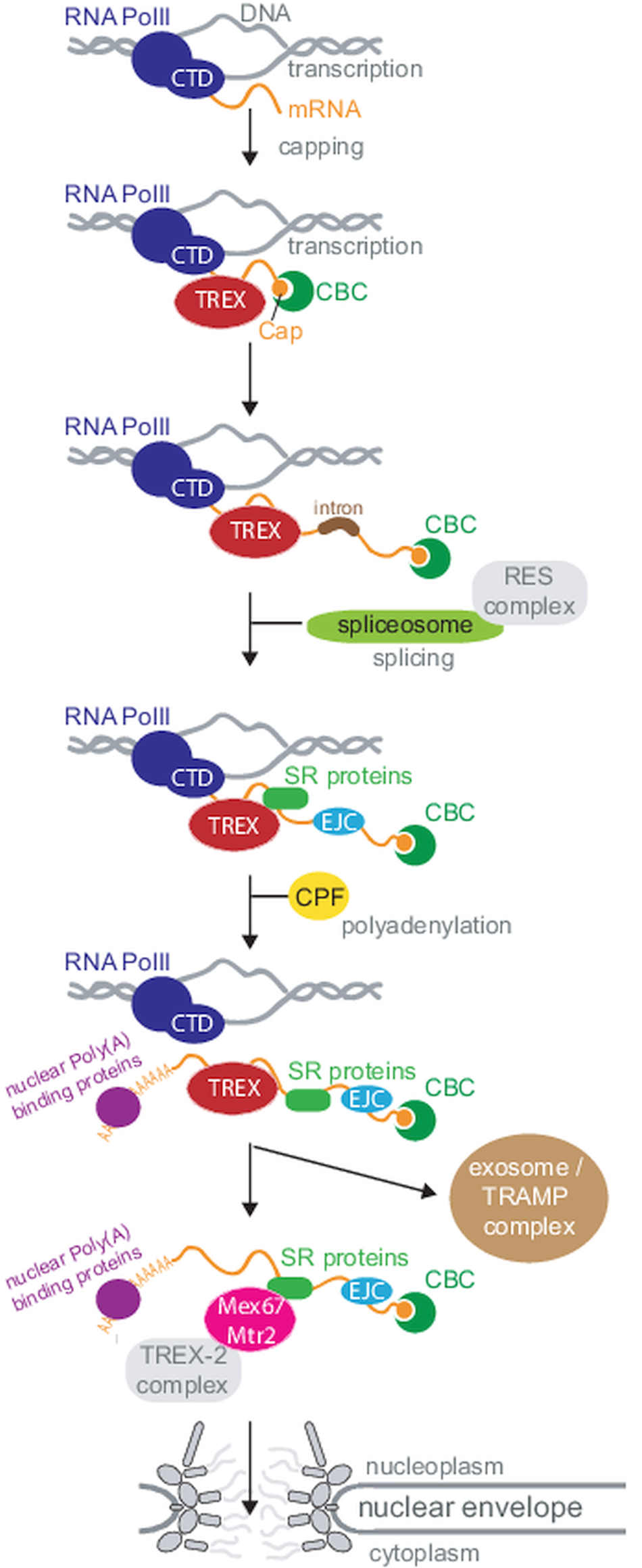
Nuclear Mrna Maturation And Mrna Export Control From Trypanosomes To Opisthokonts Parasitology Cambridge Core
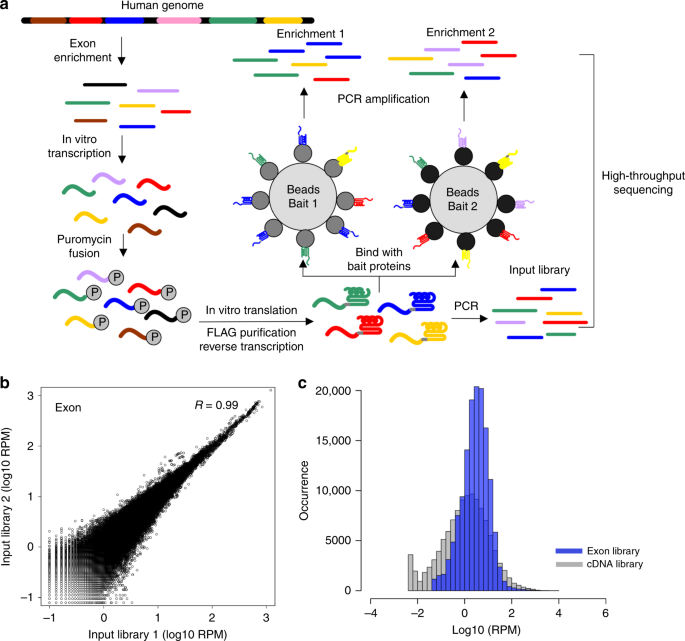
Mrna Display With Library Of Even Distribution Reveals Cellular Interactors Of Influenza Virus Ns1 Nature Communications

Artificial Multienzyme Scaffolds Pursuing In Vitro Substrate Channeling With An Overview Of Current Progress Acs Catalysis

Chemo Enzymatic Treatment Of Rna To Facilitate Analyses Muthmann 2020 Wires Rna Wiley Online Library
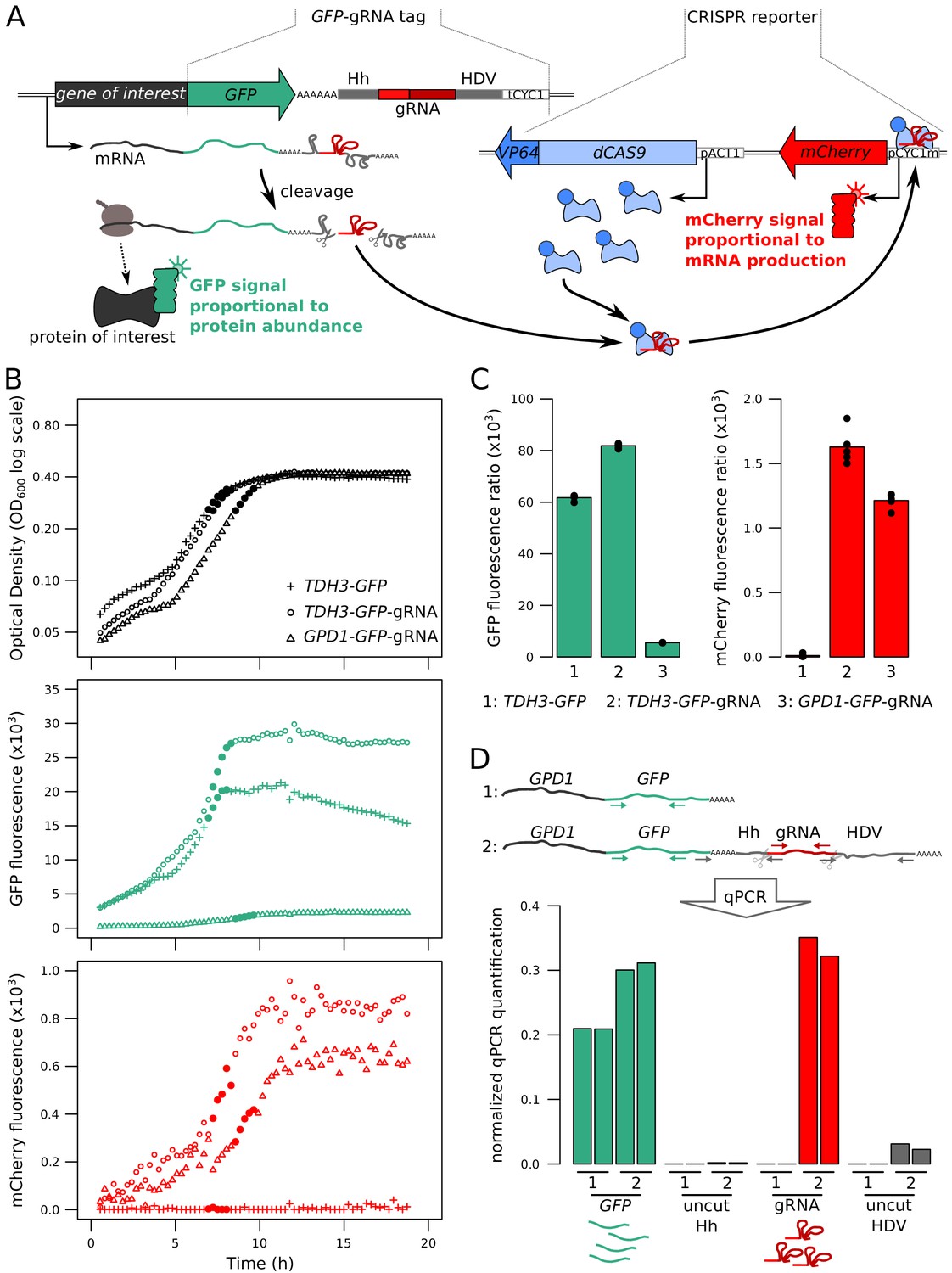
Simultaneous Quantification Of Mrna And Protein In Single Cells Reveals Post Transcriptional Effects Of Genetic Variation Elife

In Vivo Stable Isotope Labeling Of Fruit Flies Reveals Post Transcriptional Regulation In The Maternal To Zygotic Transition Molecular Cellular Proteomics













0 Response to "42 label each structure in the following diagram of mrna processing. not all labels will be used."
Post a Comment